High-throughput protein
biophysics and design
What we doProteins are gigantic molecules that perform nearly all of life's essential functions: they break down your food, replicate your DNA, generate force for muscle contraction, capture light energy in photosynthesis, and identify viruses and other invaders in your body.
We are in the middle of a revolution in computational protein design — the science of creating new proteins for unmet needs in health care, materials, and energy. Still, huge goals remain out of reach, because we have so much more to learn about the physics of proteins. Our lab develops new tools to study samples of thousands of different proteins mixed together. These large-scale experiments help us build a general understanding of protein physics, and to create more accurate, predictive computational models for protein design. We also focus on designing improved protein therapeutics. By combining computational design with large-scale experiments, we can push beyond the limits of the proteins found in nature. |
Scientifically speakingNew DNA synthesis technology now makes it possible to design and express hundreds of thousands of fully customized protein sequences, all in a mixed pool. This has revolutionary consequences for protein biophysics and protein design. We can now move beyond studying individual proteins, and instead quantify the general principles underlying protein biophysics.
To take advantage of these technologies, we develop tools that can provide high-resolution, quantitative information about entire protein libraries. We then integrate these data into new predictive models for protein design, with a special focus on designing protein therapeutics. We use Rosetta for protein design, and are members of the international Rosetta Commons. Our major tools at present are mass spectrometry proteomics, yeast and cDNA display-based selections, and deep sequencing. These tools become more powerful every year, but newer technology is always around the corner! |
Projects
Understanding protein energy landscapesAll proteins dynamically sample diverse folded, unfolded, and excited states with differing free energies. However, these ensembles of states are hidden from most experiments and have never been studied in high-throughput. We developed a new experimental approach to rapidly characterize hundreds of protein energy landscapes simultaneously using pooled protein expression and hydrogen exchange mass spectrometry. Using this approach, we plan to quantitatively define how protein energy landscapes arise from sequence and structure.
|
Design rules for hyperstable proteinsMost proteins are only moderately stable, and current generation protein therapeutics degrade and form dangerous aggregates if not stored under controlled conditions. By contrast, many designed proteins are hyperstable, maintaining their designed structures even under extreme conditions of temperature and denaturant. We are using mass spectrometry proteomics to study libraries of thousands of de novo designed proteins. This work will lead to design rules for protein therapeutics that resist degradation and aggregation under a variety of stresses, including in the digestive tract.
|
Design rules for cytosolic deliveryProtein therapeutics have limited access to the cytosol, making it difficult to design drugs that target the numerous intracellular protein-protein interactions (PPIs) critical to disease. Despite the many tools under investigation to address these challenges (each with advantages and disadvantages), there has been little clinical success. We are pursuing a new approach by engineering miniproteins to enter into cells. By using proteomics to perform unbiased screens on thousands of designs, we will build a comprehensive understanding of the determinants of cytosolic delivery, opening up an entire category of therapeutic targets to attack by new protein drugs.
|
The Team
|
PI: Gabriel Rocklin (CV) @grocklin
Postdoc: University of Washington (David Baker) Ph.D. UCSF Biophysics (Brian Shoichet & Ken Dill) B.A. Claremont McKenna College, Biology-Chemistry & History I am excited about building a great team, working with great collaborators, learning from other scientists, and trying to build a future where protein biophysics and protein design are high-throughput! |
|
Cydney Martell
Ph.D. Student, DGP Chemistry of Life Processes Training Program Fellow PhRMA Foundation Predoctoral Fellow B.A. Kalamazoo College, Chemistry with minor in Anthropology and Sociology I joined the Rocklin lab in 2020 and am interested in studying the determinants of stress-induced protein aggregation. I am using a high-throughput approach to build a model for predicting a protein’s susceptibility to aggregation. This can be used to engineer more stable proteins for therapeutic and industry applications. Outside of lab, I enjoy golfing, knitting, and cooking. |
|
Ted Litberg
Postdoctoral Scholar Ph.D. University of Denver, Biochemistry (Scott Horowitz) B.S. & M.S. Northern Illinois University, organic chemistry (Marc. J. Adler and James Horn) I’m using my expertise in uncovering mechanisms that drive the chaperone activity of nucleic acids to expand the utility of the cDNA display system. My interests outside of lab include playing volleyball, video games, cooking, and being outdoors. |
|
Andra Campbell
Ph.D. Student, DGP Biotechnology Training Program Trainee B.S. University of Michigan, Biochemistry I am interested in the structure and design of cell penetrating miniproteins (CPPs) as potential therapeutics and scientific tools. My current project includes designing a high-throughput assay to characterize the properties that allow these miniproteins to cross cell membranes. Outside of lab, I love hiking, reading, and eating my way across Chicago. |
|
Will Corcoran
Ph.D. Student, IBiS, Co-advised by Josh Leonard Biotechnology Training Program Trainee B.S. Carleton College, Biology I am investigating the use of miniproteins as binding domains for synthetic receptors, to hopefully further the capabilities of cell-based therapeutics. Outside of lab, I enjoy being outside. I love to backpack, canoe, fish, run, and try not to tear anything while playing soccer. |
|
Claire Phoumyvong
Ph.D. Student, DGP Synthetic Biology Across Scales NRT Trainee NIH Ruth L. Kirschstein National Research Service Award F31 Fellow M.B. University of Pennsylvania, Biotechnology B.A. University of Pennsylvania, Biology I am interested in the design and biophysics of cyclic miniproteins for potential therapeutics. My research includes high-throughput approaches to characterize cyclic miniprotein stability and binding affinity. Outside of lab, I love playing guitar and binge watching TV. |
|
Mario Garcia
Ph.D. Student, DGP Molecular Biophysics Training Program Trainee B.S. University of Texas at El Paso, Biochemistry I am interested in investigating the protein energy landscapes of nanobodies. My current project involves applying a high throughput approach that uses hydrogen-deuterium exchange mass spectrometry to determine the folding stability and conformational dynamics of nanobodies. This analysis could be used to develop nanobodies as therapeutics, diagnostics, and research tools. Outside of the lab, I enjoy exploring coffee shops, reading, and watching sci-fi movies. |
|
Tanu Priya
Ph.D. Student, DGP Biotechnology Training Program Cluster Fellow B.S. University of Washington, Materials Science and Engineering I am interested in learning about sequence-function relationships of enzymes using high-throughput approaches for protein characterization. I will then use machine learning to capture enzyme stability and activity into quantifiable models. The learnings from this project will help us design highly stable and active enzymes for applications ranging from chemical and pharmaceutical biosynthesis, food production, waste degradation, and regenerative medicine. Outside of the lab, I enjoy reading and exploring Chicago's food scene. |
|
Carlos Merlos
Rosetta Commons Post-Baccalaureate Scholar B.S University of Illinois Chicago, Biology I am interested in the sequence-function relationship of enzymes through the design of novel SpyCatcher structures using RFDiffusion and Protein MPNN. The insights from this project will help us to understand what makes a good enzyme in its potential application to various industries. |
|
Jane Thibeault
Postdoctoral Scholar B.S. & Ph.D. Rensselaer Polytechnic Institute, Biochemistry and Biophysics (Wilfredo Colón) I am working on incorporating my background in protein kinetic stability and proteomic mass spectrometry into the Rocklin lab’s high-throughput protein stability hydrogen-deuterium exchange (HDX) mass spectrometry assay. My focus involves peptide and point mutation localizations within the protein libraries of the HDX project. Outside of the lab, I enjoy reading, cooking, and playing music. |
|
Vasili Kalas
Physician Scientist Training Program, Division of Gastroenterology & Hepatology M.D. & Ph.D. Washington University in St. Louis, Molecular Biophysics (Scott J. Hultgren) B.S. The University of Chicago, Biochemistry I am interested in the use of computational protein binder design and high-throughput screening assays to develop novel protein-based therapeutics with application in gastrointestinal disorders. Outside of lab, I enjoy playing chess, jazz piano improvisation, traveling, and spending time with family. |
|
Állan Ferrari @ajrferrari
Postdoctoral Scholar BEPE Fellow of the São Paulo Research Foundation Ph.D University of Campinas, Science (Leandro Martínez & Fabio Gozzo) B.S. Londrina State University, Chemistry I am interested in developing high-throughput mass spectrometry-based tools to learn protein biophysics and how to use this knowledge to engineer proteins with customized features. My current project aims to apply the hydrogen-deuterium exchange mass spectrometry assay to investigate single-domain antibodies, an important protein class with broad applications in research, diagnosis and therapeutics, and which is an important step to expand the analytical pipeline to larger proteins. During my Ph.D, I worked in development of cross-linking mass spectrometry as a source of constraints to model protein-protein interactions and for protein structure prediction. In my spare time, I enjoy playing chess, cooking, hiking and hanging out with friends. |
|
Yulia Gutierrez
Undergraduate Researcher B.S. Northwestern, Biological Sciences (expected 2024) I am interested in protein engineering for therapeutic applications. My current project involves studying miniprotein aggregation in response to heat stress. Outside of lab, I enjoy baking, reading, and hiking! |
|
Pranav Doradla
Undergraduate Researcher B.S. Northwestern, Chemistry and Data Science (expected 2024) I am currently working on a project seeking to computationally identify occurrences of allosteric regulation in designed miniproteins using protease stability data. Long-term, I am interested in exploring the applications of allostery in drug design, as well as using new methods for protein design. In my free time, I enjoy singing, playing the guitar, and exercising. |
|
Elle Jung
Undergraduate Researcher B.S. Northwestern, Biological Sciences (expected 2024) I am interested in synthetic biology, specifically designing proteins that can penetrate the cell membrane in order to be used for medical applications. My hobbies include painting, cooking, and fishing. |
Lab Alumni
Jordan Gewing-Mullins, Summer Researcher 2020. Returned to Scripps College, later SynBio Apprentice, Octant Bio
Nahtalee Lomeli, Summer Researcher 2020. Currently Ph.D. student, UCSF-Berkeley Bioengineering
Jonathan Chen, Masters in Biotechnology researcher 2020-2021. Currently Process Engineer II, Kite Pharma
Radhika Dalal, RosettaCommons Post-baccalaureate scholar 2020-2021. Currently Ph.D. student, UCSF Biophysics
Wes Ludwig, Research Technician 2019-2021. Currently data analyst, Monticello Associates
Will Howe, RosettaCommons REU Summer Researcher 2021. Currently Ph.D. student, Biophysics & Quantitative Biology, UIUC
Matthew Jin, Northwestern SynBREU Summer Researcher 2021. Returned to UIUC
Kyrollos Shenouda, Masters in Biotechnology researcher 2021. Currently Ph.D. student, Bioengineering, UIUC
Andres Lira, RosettaCommons Post-baccalaureate scholar 2021-2022. Currently Ph.D. student, University of Wisconsin-Madison, Biophysics
Sarah Fahlberg, RosettaCommons REU Summer Researcher 2022. Returned to U-W Madison
Cassie Chrisman, Northwestern SynBREU Summer Researcher 2022. Currently Ph.D. student, Genomics, Bioinformatics, and Computational Biology, Virginia Tech
Kotaro Tsuboyama, Postdoctoral Fellow 2020-2023. Currently PI Tsuboyama Lab, University of Tokyo
Sugyan Dixit, Postdoctoral Fellow 2019-2023. Currently Senior Research Associate, Discovery Partners Institute
Darcy Kim, RosettaCommons REU Summer Researcher 2023. Returned to Wellesley College
Vani Lorish, Northwestern SynBREU Summer Researcher 2023. Returned to The College of New Jersey
Tae-Eun Kim, DGP Ph.D. student 2019-2023. Currently Co-Op at Abbvie.
Nahtalee Lomeli, Summer Researcher 2020. Currently Ph.D. student, UCSF-Berkeley Bioengineering
Jonathan Chen, Masters in Biotechnology researcher 2020-2021. Currently Process Engineer II, Kite Pharma
Radhika Dalal, RosettaCommons Post-baccalaureate scholar 2020-2021. Currently Ph.D. student, UCSF Biophysics
Wes Ludwig, Research Technician 2019-2021. Currently data analyst, Monticello Associates
Will Howe, RosettaCommons REU Summer Researcher 2021. Currently Ph.D. student, Biophysics & Quantitative Biology, UIUC
Matthew Jin, Northwestern SynBREU Summer Researcher 2021. Returned to UIUC
Kyrollos Shenouda, Masters in Biotechnology researcher 2021. Currently Ph.D. student, Bioengineering, UIUC
Andres Lira, RosettaCommons Post-baccalaureate scholar 2021-2022. Currently Ph.D. student, University of Wisconsin-Madison, Biophysics
Sarah Fahlberg, RosettaCommons REU Summer Researcher 2022. Returned to U-W Madison
Cassie Chrisman, Northwestern SynBREU Summer Researcher 2022. Currently Ph.D. student, Genomics, Bioinformatics, and Computational Biology, Virginia Tech
Kotaro Tsuboyama, Postdoctoral Fellow 2020-2023. Currently PI Tsuboyama Lab, University of Tokyo
Sugyan Dixit, Postdoctoral Fellow 2019-2023. Currently Senior Research Associate, Discovery Partners Institute
Darcy Kim, RosettaCommons REU Summer Researcher 2023. Returned to Wellesley College
Vani Lorish, Northwestern SynBREU Summer Researcher 2023. Returned to The College of New Jersey
Tae-Eun Kim, DGP Ph.D. student 2019-2023. Currently Co-Op at Abbvie.
We're hiring!
We are looking for undergraduate researchers, research technicians, graduate student researchers, and postdoctoral fellows. Our work is interdisciplinary, and we are looking for people with many different backgrounds including biology, chemistry, physics, and computer science.
Undergraduate research positions: Please send me a brief description of your previous experience (OK if this is primarily classes) and an explanation of why you're interested in working with us.
Research technician positions: Please send me your resume or CV, along with an explanation of what you're looking for out of your experience.
Master's student positions: We take research students from Northwestern's Master of Science in Biotechnology Program (MBP). Please get in touch if you are interested in doing your research with us!
Ph.D. student positions: If you a new Ph. D. student here, please get in touch and let's talk about your interests! For admission, apply to the Northwestern DGP program. The DGP program encompasses the entire medical school. Along with our lab, there are several other Center for Synthetic Biology labs at the medical school, including the Prindle Lab, Goyal Lab, Kelley Lab, and Choi Lab. The DGP program is also very flexible, and DGP students can rotate in labs on both the Chicago and Evanston campuses. Although our lab is sometimes able to take Ph. D. and rotation students from other programs at Northwestern, we have occasionally had difficulty convincing Evanston-based programs to permit rotations on our Chicago campus. Also, Evanston-based programs such as IBiS prefer to admit students who are primarily interested in the core faculty of those departments. Students who are primarily interested in our lab are disfavored for admission to the Evanston-based programs. If our lab is one of your main interests, I recommend applying to the DGP - a large, flexible program where you can rotate in labs on either campus.
Postdoctoral fellow positions: Please send me your CV, a cover letter explaining your goals and why you're interested in the lab, and names of three references. For more specific advice on how to apply for a postdoc position, read this thread.
Undergraduate research positions: Please send me a brief description of your previous experience (OK if this is primarily classes) and an explanation of why you're interested in working with us.
Research technician positions: Please send me your resume or CV, along with an explanation of what you're looking for out of your experience.
Master's student positions: We take research students from Northwestern's Master of Science in Biotechnology Program (MBP). Please get in touch if you are interested in doing your research with us!
Ph.D. student positions: If you a new Ph. D. student here, please get in touch and let's talk about your interests! For admission, apply to the Northwestern DGP program. The DGP program encompasses the entire medical school. Along with our lab, there are several other Center for Synthetic Biology labs at the medical school, including the Prindle Lab, Goyal Lab, Kelley Lab, and Choi Lab. The DGP program is also very flexible, and DGP students can rotate in labs on both the Chicago and Evanston campuses. Although our lab is sometimes able to take Ph. D. and rotation students from other programs at Northwestern, we have occasionally had difficulty convincing Evanston-based programs to permit rotations on our Chicago campus. Also, Evanston-based programs such as IBiS prefer to admit students who are primarily interested in the core faculty of those departments. Students who are primarily interested in our lab are disfavored for admission to the Evanston-based programs. If our lab is one of your main interests, I recommend applying to the DGP - a large, flexible program where you can rotate in labs on either campus.
Postdoctoral fellow positions: Please send me your CV, a cover letter explaining your goals and why you're interested in the lab, and names of three references. For more specific advice on how to apply for a postdoc position, read this thread.
Why join the Rocklin Lab?
Help launch a new lab. As part of a new lab, you'll be part of a tight-knit group receiving hands-on mentoring. Your success is critical to the success of the lab! You'll also help us in one of our most important endeavors - creating a positive, collaborative culture. Lab cultures are critically important and hard to change, and we want to get this right from the start.
Work collaboratively across disciplines. Our projects span cell biology, protein biophysics, computational protein design, and analysis of large datasets. Most scientific backgrounds are appropriate for the lab, including biology, chemistry, physics, and computer science. You will have the opportunity to learn both experimental technique and computational design and analysis, although you could also choose to focus exclusively on computational or experimental work. You will use your background to teach the other members of the lab, and you will learn from your peers who bring different expertise to the lab.
Small lab, big community. We are a new lab, but we are integrated into several larger communities that expand our capabilities and inspire us with their work. These communities also form an important network for our researchers, both during and after their time in the lab.
Work collaboratively across disciplines. Our projects span cell biology, protein biophysics, computational protein design, and analysis of large datasets. Most scientific backgrounds are appropriate for the lab, including biology, chemistry, physics, and computer science. You will have the opportunity to learn both experimental technique and computational design and analysis, although you could also choose to focus exclusively on computational or experimental work. You will use your background to teach the other members of the lab, and you will learn from your peers who bring different expertise to the lab.
Small lab, big community. We are a new lab, but we are integrated into several larger communities that expand our capabilities and inspire us with their work. These communities also form an important network for our researchers, both during and after their time in the lab.
- We are part of the Northwestern Center for Synthetic Biology, a highly collaborative group of labs across both Northwestern campuses, led by principal investigators at all career stages. Northwestern students also organize the GeneMods Synthetic Biology Club, with regular group discussions and invited speakers. These students are amazing!
- We are also part of the RosettaCommons, an international collection of over 400 researchers focused on biomolecular modeling and design using Rosetta. This community is an incredible resource for our research, and for building friendships within science. Rosetta researchers congregate every year at RosettaCon in Leavenworth, Washington. Expertise with Rosetta is highly sought-after in the biotech industry and in academia.
- We make extensive use of proteomics, and are part of the strong Northwestern Proteomics community. Northwestern is a world-leader in Top Down Proteomics, which is an ideal approach for examining our computationally designed libraries of proteins.
Papers
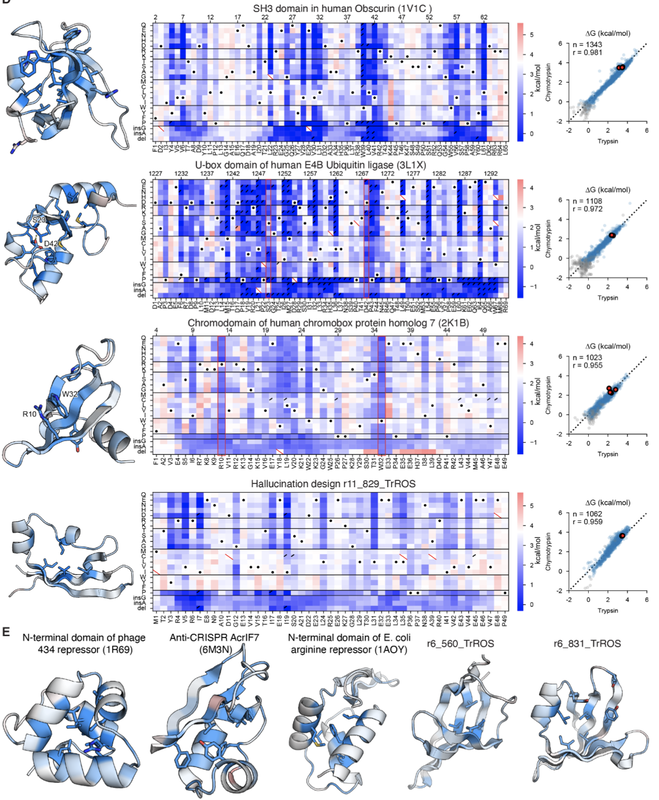
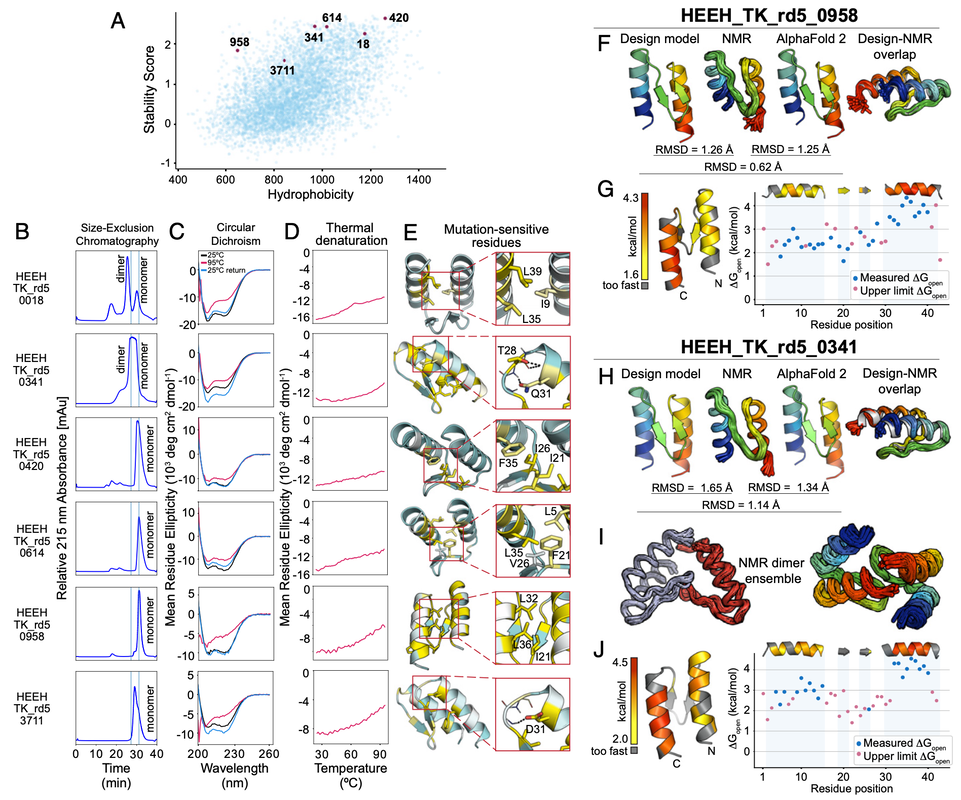
Protein folding stability goes MEGA-SCALE!! Now out in Nature!Mega-scale experimental analysis of protein folding stability in biology and design
Tsuboyama et al. Nature 2023 All data available on Zenodo! Please register your use of the data so that we can continue to generate more! Nature Research Briefing (short summary) Tweet threads from Gabe and Kotaro! 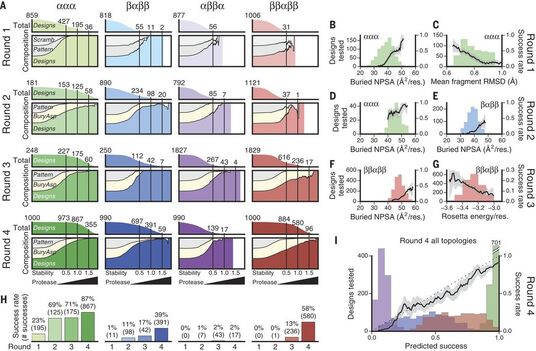
Global analysis of protein folding using massively parallel design, synthesis, and testing
Rocklin et al. 2017 Perspective by Woolfson et al. Research highlights: Nature Chemical Biology, Nature Methods News coverage: Chemical & Engineering News (with cover art), Chemistry World, Genetic Engineering & Biotechnology News, The Scientist |
Dissecting the stability determinants of a challenging de novo protein fold using massively parallel design and experimentation
Kim*, Tsuboyama* et al. PNAS 2022 BioRxiv Open Access Link Tweet threads from Gabe and Kotaro! 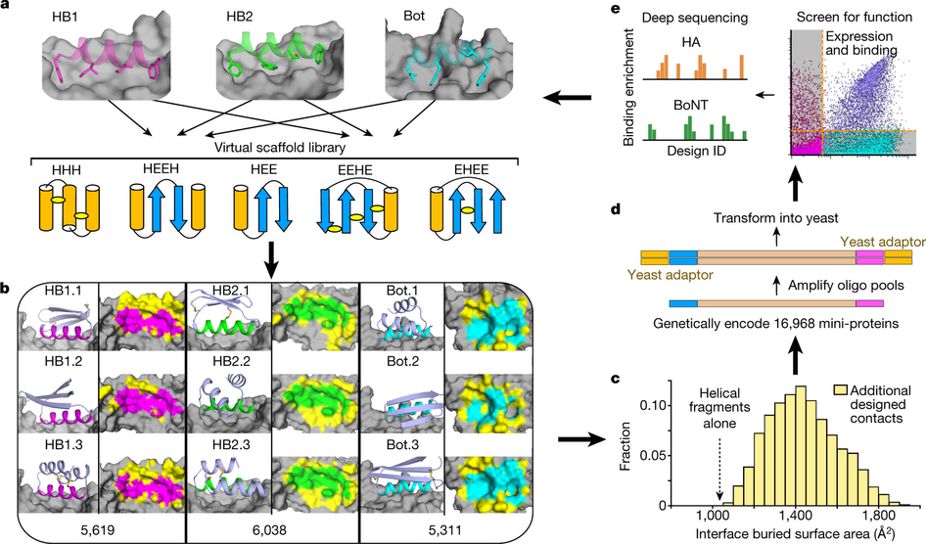
Massively parallel de novo protein design for targeted therapeutics
Chevalier*, Silva*, Rocklin* et al. 2017 Research highlights: Cell, Biochemistry News coverage: The New York Times, Chemical & Engineering News, In The Pipeline |
Full publication list on Google Scholar
Contact
|
Gabriel Rocklin
grocklin@gmail.com Twitter: @grocklin 303 E Superior St Simpson Querrey building 11-517 Chicago, IL 60611 Northwestern Feinberg School of Medicine Department of Pharmacology Center for Synthetic Biology Chemistry of Life Processes Institute Robert H. Lurie Comprehensive Cancer Center |